For examining one or more genomes I describe an easy procedure using some free software packages from the Net:
1- Download the genomes from "The NCBI ftp site " or from other Web site.
2- Go to genome and choise the folder : example Virus.
3- Open the folder and select your genome and store it in your computer.
4- Select only genome file with end: .gbk or .gb or.ffn or Fasta.
5- Download :
BugView (genome browser for comparing the arrangement of genes on a pair of related genomes, and can also be used to view individual genomes).
DAMBE (software package for extensive data analysis in molecular biology and evolution).
e-Workbench (for comparative genome analysis).
(read all user manuals but they are simple and intuitive ):
6- Download the genome file by DAMBE and by e-Workbench.
7- Read by e-Workbench, one by one, the genome features and also check them by DAMBE.
8- Write by Excel programme each information regarding the genes.
9- Repeat points 6,7,8 for another genome.
10- Open BugView and compare two genomes.
Example: D29 and L5 mycobacteriophages
These are the results of my genome analysis using the procedure that I have described above:
1- This is the Excel table where I compare D29 genome against L5 genome.
2- This is the Excel table where I compare L5 genes against D29 genes.
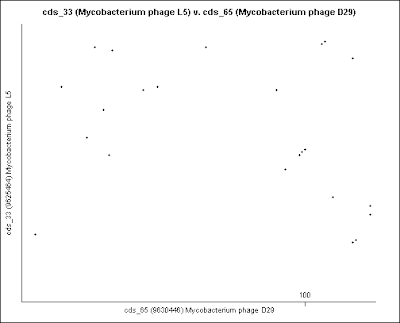
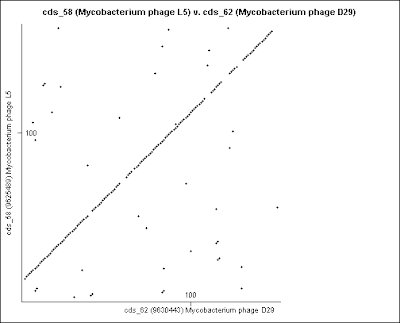
3- This the Gene comparison image between L5 and D29 genes from the Excel table ( point 2).

4- These are the Dot plot images for Gene comparison, in numerical order, from the Excel table (point 2).











These genomes are not circular. They are opened in the circular viewer.


Genome comparison between D29 and L5 mycobacteriophages by BugView.

Genomic skew in D29 and L5 mycobacteriophages
The frequencies of bases in the genome of an organism are not always equiprobable. For example, the composition can have high "GC" content relative to the "AT".
The cause of 'skew' is not understood. Some possibilities include strong biases in mutation or DNA repair.
The minimum and maximum of a GC-skew can be used to predict the origin of replication (minimum) and the terminus location (maximum) in procaryotic genomes. Since the coding strand of bacterial genomic DNA tends to be purine rich and majority of genes are transcribed in the same direction as the movement of the replication fork there is asymmetric nucleotide composition along the
genome such that the DNA composition may
be used to predict the origin and termini of replication.
The origin of replication (ori) and replication terminus (ter) can be deduced by GC skew and cumulative GC skew analyses.

Purine skews are calculated from the first position in the sequence to the last: for each nucleotide, increment a counter if this nucleotide is a purine; decrement it if it is a pyrimidine. The effect is to compute the number of purines minus the number of pyrimidines from the first position to the current one. The X-axis of the skew graph is the position in the sequence; the Y-axis is the value of the counter at this position.

Keto and dinucleotide skews are calculated analogously, with the obvious differences.For a window size of k, every k'th position is drawn

The cumulative dinucleotide skews display the abundance of one nucleotide relative to another across the length of a DNA sequence that may represent a single gene or a complete genome.GC and AT skews have been widely used to predict termini and origins of replication in bacterial and mammalian genomes,
transcription start sites in plants and fungi ,
as well as transcription regions in the human genome .




The "DNA walk" is another method used to study
nucleotide distribution, first described by Lobry and used to detect origins of DNA replication in bacteria genomes.
To graph a DNA walk, a direction (North, South, East, and West) is assigned to each of the four nucleotides and the sequence is then plotted on a graph, beginning at (0,0) and moving one step in the direction specified by each successive nucleotide.

